Enzymes


Please check back later for additional changes
An enzyme is a protein that catalyzes a chemical reaction, greatly speeding it up while not being consumed by the reaction. This allows enzymes to be active even in very low concentrations. Enzymes play an important role in the creation of all fermented beverages, and more generally, they are needed for all life processes.[1] As with all proteins, enzymes have particular temperature and pH ranges in which they function, and more narrow ranges in which the activity is considered optimal. The effect of temperature is greater than the effect of pH. Knowing the optimal ranges can be helpful, but it must be realized that the enzymes will be active to some extent outside those ranges.[2] Enzymes denature (the three-dimensional structure unfolds) at higher temperatures, rendering them inactive.[1] Enzymes tend to have a very specific substrate upon which they act, and therefore are often named after the substrate, adding "-ase" to the end.[3]
Coenzymes: The action of many enzymes is tied to the presence of an additional non-protein component that binds with its structure. For example, bivalent metal ions (e.g. iron, magnesium, calcium) are often involved as coenzymes.
Isoenzymes: Enzymes that have different structures but catalyze the same reaction are called isoenzymes. Each isoenzyme may have different characteristics such as optimal temperature and pH ranges. Generally, most enzymes in living organisms have several isoenzymes.
Natural enzymes
Malting
Coming eventually
Enzymes are of special importance during malting. In post-harvest maturity, harvested malting barley contains a huge amount of enzymes and their precursors. From the malting point of view, enzymes of the hydrolasis group can be classified as the most important enzymes. They can be divided into four groups (cytolytic enzymes, proteolytic enzymes, phosphatases, amylases) and the class of oxidoreductases, followed then by transferases, lyases, isomerases, and ligases.[4]
- Cytolytic enzymes
Mashing
During mashing, a very large number of enzymes act simultaneously on the components of the grist under conditions that are far from optimal for many of them in terms of substrate concentration and accessibility, pH, and enzyme stability. Enzymes are progressively inactivated at different rates depending on the temperature, the pH, the presence of substrate and other substances (such as tannins and cofactors such as calcium ions) in solution.[2] Starch, proteins, nucleic acids, lipids and other substances are degraded, usually by hydrolytic (cleaving) reactions, but other reactions, such as oxidations, also occur.[5]
- Starch and sugar degradation (see Starch and Sugars)
- α-amylase (optimal 72–75°C, pH 5.6–5.8) degrades starch and dextrins into smaller sugars by cleaving α-1,4-bonds.[6][7][1][2] Rapid inactivation occurs at 78–80°C and above.[8][7]
- β-amylase (optimal 60–65°C, pH 5.4–5.6) releases maltose from the ends of sugar chains by cleaving α-1,4-bonds.[6][7][1] Rapid inactivation occurs at temperatures of 65-70°C and above.[8][7][9]
- Limit dextrinase AKA debranching enzyme AKA R-enzyme AKA pullulanase (optimal 55–65°C, pH 5.4) degrades limit dextrins into dextrins/sugars by cleaving the branch point (α-1,6 bonds).[6][7][10] Inactivation occurs at temperatures of 65-75°C and above, although it's not destroyed unless boiled.[8][7][9]
- α-glucosidase AKA maltase (optimum 35–45°C, pH 4.6–6.0) degrades maltose, isomaltose, oligosaccharides, dextrins and starch, cleaving single glucose units from the ends of chains (mainly α-1,4 bonds, but also some α-1,6 bonds).[2][6][11][12][13] This class of enzyme has not been studied to the same extent as the other starch-degrading enzymes.[14]
- Glucoamylase (optimal 35–40°C) cleaves a single glucose unit from the end of any sugar chain (both α-1,4 and α-1,6 bonds).[15][11] Its activity is virtually non-existent during mashing because of its very low optimal temperature.
- Invertase (optimal 50°C, pH 5.5) splits sucrose into glucose and fructose. Active up to 62–67°C.[7]
- Protein degradation (see Protein)
- Endopeptidases, which include metalloproteases, cysteine proteases, aspartic proteases, and serine proteases (optimal 45–50°C, pH 3.9–5.5) over 40 different endopeptidase enzymes degrade proteins into peptides and free amino acids.[6][1]
- Carboxypeptidases (optimal 50°C, pH 4.8–5.6) degrade proteins & peptides into free amino acids.[6][1]
- Aminopeptidases (optimal 45°C, pH 7.0–7.2) degrade proteins & peptides into free amino acids.[6][1] Inactive during mashing.
- Dipeptidase (optimal 45°C, pH 8.8) degrades dipeptides into free amino acids.[6][1] Inactive during mashing.
- Beta-glucan liberation and degradation (see Beta-glucans and arabinoxylans)
- β-glucan solubilase (optimal 62–65°C, pH 6.8) releases high-molecular-weight matrix-bound β-glucans, increasing the amount in the wort.[6][16]
- Endo-(1,3;1,4)-β-glucanase (optimal 48°C, pH 4.7) degrades soluble high-molecular-weight β-glucan into low-molecular-weight β-glucan.[6][17]
- Endo-(1,3)-β-glucanase degrades soluble high-molecular-weight β-glucan into low-molecular-weight β-glucan, and may also help solubilize β-glucan.[18][19]
- Endo-(1,4)-β-glucanase AKA cellulase degrades soluble high-molecular-weight β-glucan (including cellulose) into low-molecular-weight β-glucan.[18][19][20]
- Exo-β-glucanase (optimal <40°C pH 4.5) degrades glucose from the ends of β-glucan.[6][19]
- Phosphate liberation (see Phosphates)
- Lipid degradation and oxidation (see Lipids)
- Lipase (optimal 55–65°C, pH 6.8–7.0) degrades lipids & lipid hydroperoxides into glycerine plus free fatty acids, and/or hydroperoxides.[6][21][22][20]
- Lipoxygenases (optimal 45–55°C, pH 6.3–7.0) oxidizes fatty acids into fatty acids hydroperoxides.[6][21][20]
- Hydroperoxide lyase transforms lipid hydroperoxides through a series of steps into staling compounds such as trans-2-nonenal.[20]
- Phenolic compound release or oxidation (see Phenolic compounds, Oxidation)
- Polyphenol oxidase (optimal 60–65°C, pH 6.5–7.0) oxidizes polyphenols.[6]
- Feruloyl esterase AKA ferulic acid esterase AKA cinnamoyl esterase (optimal activity 40–50°C, pH 5.2–6.6) liberates phenolic acids (mainly ferulic acid) from arabinoxylans.[7][23] Inactive at 65°C and above.[24]
- Non-specific oxidation (see Oxidation)
- Other
- Endo-xylanase, exo-xylanase, and arabinosidases (optimal 45°C) degrade pentosans.[7]
- Pentosan solubilase releases bound pentosans.[7]
- Phosphorylase cleaves the terminal alpha-(1, 4) links in non-reducing chain ends with inorganic phosphate to release glucose-1-phosphate. Apparently its possible role in mashing has never been investigated.[2]
- Catalase catalyses the conversion of peroxides to water and ground state (unreactive) oxygen, however it is rapidly destroyed during mashing at 149°F (65°C) and therefore it is largely irrelevant in the brewhouse.[25]
- Superoxide dismutase catalyses the formation of peroxides from superoxides which in the absence of catalase leads to the formation of the hydroxyl radical.[25]
Fermentation
Coming eventually
- Yeast enzymes
- Invertase breaks sucrose into its constituents glucose and fructose.
Wine
Coming eventually
Added enzymes
A variety of enzyme products ("exogenous" enzymes) are available to home brewers for various purposes such as increasing starch/dextrin degradation or decreasing haze.
General information:
Storage - Enzyme preparations are not stable, so they should be stored cool and used fresh since the activity decreases over time.[2][26] Refer to the manufacturer for specific recommendations.
Impurities - Enzyme products are created by living organisms.[20] Therefore these products are not "pure", and will usually contain other substances such as residual materials from the nutrient medium in which the microbes were cultured, other enzymes besides the one(s) specified, diluents, extenders or carriers, and preservatives. Be aware that these impurities can potentially lead to haze formation, deterioration of beer foam, and loss of yeast flocculation. Modern enzyme products usually do NOT contain viable microbes.[2][26]
Product comparison - Products from different suppliers should be considered distinct, and it can be difficult to make comparisons between them due to lack of standardization. Also, the action of enzyme products can be greatly influenced by the usage conditions (e.g. temperature and pH), and therefore the results may vary between different brews. Consequently, the effectiveness of the addition of an enzyme preparation must be determined by brewers under their particular processing conditions.[2]
Common products:
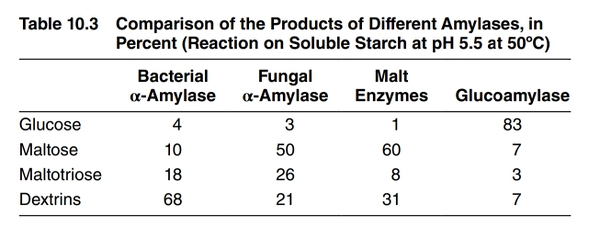
- Amylase (bacterial) - Degrades starch to dextrins very effectively but does not produce much fermentable sugar. Used mainly for liquefaction of adjunct starch.[26] See Bacterial alpha amylase.
- Fungal Alpha Amylase - Degrades larger dextrins, producing limit dextrins plus some fermentable sugars. Used to mildly increase fermentability. See Fungal alpha amylase.
- Glucoamylase - Degrades dextrins to produce mostly glucose, greatly increasing fermentability. Used to produce dry beer (e.g. "Brut IPA"), low-carb beer, or to accelerate sour beer production. See Glucoamylase.
- Clarity Ferm AKA Clearzyme AKA Clarex? is a proline-specific endoprotease that reduces chill haze and reduces "gluten"[20]
- Glucabuster
- Lysovin (lysozyme)
- Alpha galactosidase from AIH
See also
Potential sources
- http://lowoxygenbrewing.com/forum/viewtopic.php?f=11&t=1821
- Amino Acid Permeases and their Influence on Flavour Compounds in Beer
- https://scholar.google.com/scholar?hl=en&as_sdt=1%2C36&q=sun+A+quantitative+assessment+of+the+importance+of+barley+seed+alpha-amylase%2C+beta-amylase%2C+debranching+enzyme+and+alpha-glucosidase+in+starch+degradation.&btnG=#d=gs_qabs&u=%23p%3DRr5Dbt7Zj7MJ
- https://www.researchgate.net/profile/Ahmed_Gomaa35/publication/323252887_Application_of_Enzymes_in_Brewing/links/5b5f33ae458515c4b2531f59/Application-of-Enzymes-in-Brewing.pdf
- https://hibernianbrewingschool.ie/wp-content/uploads/2015/09/The-role-of-enzymes-IOB.pdf
- http://www.knudsenbeverageconsulting.com/wp-content/uploads/2011/mbaa/mbaarmdpresentationenzymesinbrewing51102.pdf
- http://themodernbrewhouse.com/forum/viewtopic.php?f=11&t=2168
- https://www.themodernbrewhouse.com//forum/download/file.php?id=1931
- https://prowm.com/wp-content/uploads/2019/06/PRO-Tech-Notes-ISSUE-6-VOLUME-2.pdf
- https://www.researchgate.net/profile/Ahmed_Gomaa35/publication/323252887_Application_of_Enzymes_in_Brewing/links/5b5f33ae458515c4b2531f59/Application-of-Enzymes-in-Brewing.pdf
- Scheffler, A. and Bamforth, C.W. (2005) Exogenous β-glucanases and pentosanases and their impact on mashing, Enzym. Microb. Tech. 36, 813–817.
- Bamforth, C.W. (2010) The enzymology of cell wall breakdown during malting and mashing: An overview, Tech. Q. Mast. Brew. Assoc. Am.
References
- ↑ a b c d e f g h Kunze W. Wort production. In: Hendel O, ed. Technology Brewing & Malting. 6th ed. VBL Berlin; 2019. p. 230.
- ↑ a b c d e f g h i Briggs DE, Boulton CA, Brookes PA, Stevens R. Brewing Science and Practice. Woodhead Publishing Limited and CRC Press LLC; 2004.
- ↑ a b c Fix G. Principles of Brewing Science. 2nd ed. Brewers Publications; 1999.
- ↑ Benešová K, Běláková S, Mikulíková R, Svoboda Z. Activity of proteolytic enzymes during malting and brewing. Kvasný Prům. 2017;63(1):2–7.
- ↑ Szwajgier D. Dry and wet milling of malt. A preliminary study comparing fermentable sugar, total protein, total phenolics and the ferulic acid content in non-hopped worts. J Inst Brew. 2011;117(4):569–577.
- ↑ a b c d e f g h i j k l m n o p Krottenthaler M, Back W, Zarnkow M. Wort production. In: Esslinger HM, ed. Handbook of Brewing: Processes, Technology, Markets. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2009.
- ↑ a b c d e f g h i j Narziss L, Back W, Gastl M, Zarnkow M. Abriss der Bierbrauerei. 8th ed. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2017.
- ↑ a b c Visser MJ. Evaluation of malted barley with different degrees of fermentability using the Rapid Visco Analyser (RVA). University of Stellenbosch. 2011.
- ↑ a b Evans DE, Fox GP. Comparison of diastatic power enzyme release and persistence during modified Institute of Brewing 65°C and Congress programmed mashes. J Am Soc Brew Chem. 2017;75(4):302–311.
- ↑ McCafferty CA, Jenkinson HR, Brosnan JM, Bryce JH. Limit dextrinase — Does its malt activity relate to its activity during brewing? J Inst Brew. 2004;110(4):284–296.
- ↑ a b Guerra NP, Torrado-Agrasar A, López-Macías C, et al. Use of Amylolytic Enzymes in Brewing. In: Preedy VR, ed. Beer in Health and Disease Prevention. Academic Press; 2009:113–126.
- ↑ Bamforth CW, Fox GP. Critical aspects of starch in brewing. BrewingScience. 2020;73:126–139.
- ↑ Im H, Henson CA. Characterization of high pI α-glucosidase from germinated barley seeds: substrate specificity, subsite affinities and active-site residues. Carbohydr Res. 1995;277(1):145–159.
- ↑ Andriotis VME, Saalbach G, Waugh R, Field RA, Smith AM. The Maltase Involved in Starch Metabolism in Barley Endosperm Is Encoded by a Single Gene. PLoS ONE. 2016;11(3):1–13
- ↑ Vriesekoop F, Rathband A, MacKinlay J, Bryce JH. The evolution of dextrins during the mashing and fermentation of all-malt whisky production. J Inst Brew. 2010;116(3):230–238.
- ↑ a b c Sacher B, Becker T, Narziss L. Some reflections on mashing – Part 2. Brauwelt International. 2016;6:392-397.
- ↑ Jin YL, Speers RA, Paulson AT, Stewart RJ. Barley β-glucans and their degradation during malting and brewing. Tech Q Master Brew Assoc Am. 2004;41(3):231–240.
- ↑ a b Muller R. Factors influencing the stability of barley malt β-glucanase during mashing. J Am Soc Brew Chem. 1995;53(3):136–140.
- ↑ a b c Kanauchi M, Bamforth CW. The relevance of different enzymes for the hydrolysis of β-glucans in malting and mashing. J Inst Brew. 2008;114(3);224–229.
- ↑ a b c d e f Evans E. Mashing. American Society of Brewing Chemists and Master Brewers Association of the Americas; 2021.
- ↑ a b Golston AM. The impact of barley lipids on the brewing process and final beer quality: A mini-review. Tech Q Master Brew Assoc Am. 2021;58(1):43–51.
- ↑ Schwarz P, Stanley P, Solberg S. Activity of lipase during mashing. J Am Soc Brew Chem. 2002;60(3):107–109.
- ↑ Schwarz KJ, Boitz LI, Methner FJ. Release of phenolic acids and amino acids during mashing dependent on temperature, pH, time, and raw materials. J Am Soc Brew Chem. 2012;70(4):290–295.
- ↑ Wannenmacher J, Gastl M, Becker T. Phenolic substances in beer: Structural diversity, reactive potential and relevance for brewing process and beer quality. Compr Rev Food Sci Food Saf. 2018;17(4):953–988.
- ↑ a b EtokAkpan OU. Preliminary study of fat oxidation in sorghum and maize brewing. World J Microbiol Biotechnol. 2004;20:569–573.
- ↑ a b c Ryder DS. Processing aids in brewing. In: Stewart GG, Russell I, Anstruther A, eds. Handbook of Brewing. 3rd ed. CRC Press; 2017.